Idiots look at Data, Part II: Cursory Glance
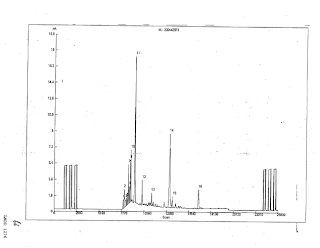
In Part I, we identified things we'd be looking for, and offered a model of "good". Now we'll do a cursory run through showing the big picture of what we're going to do. For all the samples in Ex-107's Table 3 for which we have the data, we're going to look at the F3 IRMS graph:
We should all remember that isotopic ratio has no relation to peak height. Without more ado:
[MORE]

No reported values available.
Ex 92 - April testing of 993865 - LNDD 1398 (Landis from 3-Jul). There is no reported value for the 5aA, with no explanation given. It could be a decision to say it has matrix interference. 5bA reported as -1.04
Exhibit for for 993856 from 11-Jul not available, reported as 5aA -2.91 and 5bA at -4.09
Ex 88 - April testing of 993855 - LNDD 1015 (Landis from 13-Jul). 5aA reported as -4.62, 5bA as -4.09
Ex 90 - April testing of 825425 - LNDD 1207 (Landis from 14-Jul), reported as -1.01, -0.70
Ex 86 - April testing of 825428 - LNDD 824 (Landis from 18-Jul). Reported as -5.06, -3.56
USADA 173, reported as 5aA -6.14, 5bA -2.15, Andro (other fraction) -3.99. Done on the IsoPrime1 machine with different software, so the display is different.

USADA 349, the Stage 17 B sample, reported as 5aA -6.39, 5bA -2.15, and Andro -3.51. Also done on the Isoprime1 with different display.
Ex 87 - April testing of 825429 - LNDD 922 (Landis from 22-Jul). Reported as -5.06, -3.56
Ex 84 - April testing of 825424 - LNDD 633 (Landis from 23-Jul). Reported as -4.96, -1.45.
Ex 93 - April testing of 825427 - LNDD 1492, the ocean blue, (Aguilera control), reported at 5aA -1.31, 5bA -0.95
Ex 85 - April testing of 825426 - LNDD 730 (Aguilera control), reported as 5aA -0.77, 5bA -0.88
Ex 89 - April testing of 825423- LNDD 1193 (Aguilera control), reported at 5aA -1.21, 5bA -0.79
We'll zoom into the regions of interest in following parts of this series.










10 comments:
What information IS conveyed by the peak heights in an IRMS graph?
The IRMS graphs you're showing that were produced by the IsoPrime1 machine look a lot more familiar than the other graphs - the ones identifying the peaks by peak count. Take, for example, LNDD 0894, your figure 6. What is going on here? There are 22 peaks, and there's no indication of the delta-delta for any of these peaks. I'll put aside my favorite issue of how you'd identify these peaks, and ask, how do you assign delta-delta values to these peaks?
(M, I'm reasonably certain that even OMJ would have trouble doing pattern matching on Figure 6. Sorry. I know we're not talking about peak identification at the moment. I just couldn't help myself.)
Larry,
I don't think OMJ would have any problem with figure 6. There are 4 central peaks around peaks 19 and 20 with the retention times at 1300+ seconds. These are the same 4 central peaks as in the stage 17 F3 graph also at around 1300+ seconds. There's a couple more pronounced trailing peaks here when compared to the stage 17 graph. Peak 19 is the 5B and peak 20 is the 5A. Larry you do use RRT and RT to locate your peaks,they just don't have to match within 1%.
I believe, the heights of the GC-IRMS peaks measure the amount of carbon atoms eluting at that point in time.
So have you looked at the Shackleton figure 3 yet?
In the graphs below A=5B, and B=5A. Was the 5A identified in the F3 sample. Yes.
GRAPH OF RRT RESULTS
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||scale
1) GCMS Mix Cal
................I........a.A.B.d......P
2) GCMS F3
................I........a.A.B.d......P
3) GC-IRMS F3
......................I..........a.A.B.d.......P
4) GC-IRMS F3 Blank Urine
......................I..........a.A.B.d.......P
5) GC-IRMS Mix Cal
......................I............A............
The B in 1) and 2) match based on RTs.
The metabolites in the IRMS graphs in 3), 4), 5) eluted later than in 1) because the IRMS involved a combustion phase and because changed ramp conditions caused the mebolites to elute further apart probably to achieve greater peak separation.
The RTs in the graphs in the IRMS all matched. So there is no question that the B in the IRMS F3, must be the same B as in the GCMS F3. Remember it comes right after the A, which we have fixed in the 5) graph.
M, yes I've seen the Shackleton figure 3. I do everything you tell me to do!
Where did you come up with your 9:33 graphs?
My impression is that you work much harder at peak identification than LNDD ever did! But if you want to discuss peak identification, we should probably do it on a different post, because I don't think it's exactly on topic here. (And yes, it IS my fault, I'm the one who brought it up here.)
M, are your 9:33 AM graphs based on the F3 A sample or the F3 B sample. If you put me under oath, I'd probably end up agreeing with you and OMJ about the A sample. The B sample is not as clear-cut IMHAUAAIAAIO (in my humble and unscientific, after all I am an "idiot", opinion).
Larry,
My graphs are rough scale text representations of the corresponding B sample graphs. I could have created graphs in excel with the actual values, but didn't know how to post them.
Each space or period represents around 45-40 seconds. So for example the Internal Standard elutes in the IRMS about 200+ seconds after the IS in the GCMS. The 5A came about 100 seconds longer after the IS in the IRMS when compared to the GCMS, so I added 2 spaces to that distance. The other spacings are rough approximations. So there was only 30-50 seconds between the a, A, B, d, so I placed 1 space between them.
Frankly, I don't remember any differences between the A and B samples. I may take a look back. Wait, I don't think they used a mix cal for quality control in the A sample IRMS.
M, has OMJ ever said that the right way to go is to use RRTs with a standard more relaxed than 1%? I thought he said to do pattern matching or peak counting.
As you know, I do not agree that the 1% - 12 second standard can be relaxed in this case under TD2003IDCR. However, if you're going to try to advocate such criteria, you'll probably have to add to your criteria the assumption that the metabolic peaks of interest will always appear on the graph in the same order. Some of the peaks naturally appear very close together - I think it's 5bA and 5aA. If you relax your 1% standard too much, you won't be able to distinguish one from the other ... unless you add to your criteria that one always appears before the other.
Larry,
Retention times and relative retention times are usually measured from some internal standard. The retention time and relative retention time always establish the order of elution of a peak relative to that internal standard.
The only question here is whether the slight change in GC ramp conditions, (the IRMS was at a slightly lower temperature ramp so the metabolites in theory should elute more slowly), caused a peak (5A) to EXACTLY switch positions with some other peak (not including the 5B which we have fixed). The chances of this are close to zero given all the other structural information we have in the graphs. Otherwise we know that the 5A must directly follow the 5B. Which it does in this case.
M -
Let me use a non-real world demonstration to illustrate my point about relaxing the TD2003IDCR 1% standard.
We'll make this a simple RT example without a reference internal standard, for ease of illustration, and I'm going to round my calculations to the nearest integer. Let's say that you're trying to identify 3 metabolites. On the GC/MS test, you identify metabolite 1 with a RT of 10:00 (600 seconds), metabolite 2 with an RT of 10:30 (630 seconds) and metabolite 3 with an RT of 20:00 (1200 seconds). On your IRMS graph, you have three peaks with RTs of 10:11 (611 seconds), 10:25 (625 seconds) and 20:50 (1250 seconds). Can you identify the IRMS peaks using RTs?
First thing to note is that the 1% standard isn't going to work. The first GC/MS peak is at 600 seconds, setting up a possible RT range (+/- 1%) of 594 to 606 seconds. We don't have an IRMS peak in that range.
So, you might argue, the conditions of this test justify relaxing the 1% standard to 2%. If we use a 2% standard, then we can identify peak 1 and peak 2. But peak 3 is still outside of the 2% range.
So, we grit our teeth and argue that the conditions of this test were so demonstrably bad, we could justify a 5% standard of error. That should solve our problems, you might think. And yes, the 5% standard DOES allow us to identify peak number 3. BUT ... a 5% standard causes us a BIG problem with peaks 1 and 2.
Remember, peak 1 had a GC/MS RT of 600 seconds, so with a 5% margin, we could identify peak 1 to any peak on the IRMS chart with an RT between 570 and 630 seconds. Similarly, we can identify GC/MS peak 2 to any peak on the IRMS chart with an RT (roughly) between 599 and 661 seconds. Our problem is that BOTH of the first two peaks on the IRMS chart fit into EACH of these time ranges. If you strictly apply your 5% margin of error, you can no longer tell which peak is which.
In effect, your margin of error is now so wide, it allows for the possibility that the two IRMS peaks have shifted in order from where they were shown on the GC/MS graph.
This is why I'm saying that when you apply your relaxed RT standards under your reading of TD2003IDCR, you'll probably also need to add the criteria that IRMS peaks cannot shift from the order they were shown on the GC/MS graph.
Larry,
1.To compare elutions in a GC-IRMS with those in a GCMS one must use RRTs computed from an internal standard, not RTs because they are different types of machines.
2. The 1% RRT standard only applies to chromatography performed on the same GC machine under identical ramp conditions.
2. You cannot compare RRTs in a GC-IRMS sample with RRTs of a GCMS reference material and use the 1% standard. They will never match. Meier's testimony was that even under ideal attempted identical GC ramp conditions the RTs will be off by 1 to 2 minutes implying the RRTs will be off by 2-3%.
3. Ergo, if one wants to use the TD2003IDCR RRT identification criteria with the GCMS to GC-IRMS procedure one must relax the 1% criteria.
4. Your example is wrong in that you always have to use an internal standard from which you can calculate the RRT of each peak. Using those RRTs you can establish a peak order. I also suspect that the values you use are unrealistic. In particular the 611 IRMS RT is way too close to the 600 GCMS RT because the IRMS will always have a big delay. I tried to work out a numerical example using your figures and an inserted IS RT, but the RRT error was all over the place.
5. Remember that we perform three GCs here with an internal standard inserted into each: 1) GCMS reference material, 2) GCMS sample, 3) GC-IRMS identical sample as in 2).
We know the sequence of peaks in the reference sample, but not its exact RT because that will depend on the GC ramp conditons and machine. We identify each metabolite in the GC sample by comparing its RT with the reference material RT AND each sample metabolite's mass spectrum. So we know the identity and sequence of each metabolite peak in the sample and their relative distance from each other using RRTs.
6. We then perform the GC-IRMS on the identical sample. We know that the IRMS RRTs of peaks will differ from the GCMS RRTs by a minimum of 2-3%, and in the Landis case by 4-7% as the metabolite elutes further from the IS. But we assume that their sequence will be the same as measured from the IS, and the relative distance between them will be relatively close because it's the same sample.
7. So to answer your question. Yes, I believe that the TD2003IDCR standard if/when applied to GCMS to GC-IRMS procedure assumes that the peaks will elute in the same order.
But I am still not sure that TD2003IDCR was specifically thought to apply to the GC-IRMS procedure. LOL!
Post a Comment