Identification Legal, continuing.
Picking up the conversation from comments in Larry: ISL Violations any way you look, we take the last few comments and start again.
We too would like to thank M for his diligence in posting and continuing the discussion. It wouldn't be anywhere near as revealing and interesting if he weren't around.
m said, (with clarification via email later)...
What Identification Criteria were used in the IRMS test?
1) What criteria did the lab purport to use to identify the metabolites that were tested? 2) What criteria did the lab actually use?
The short answer to 1) is that USADA Pre-Hearing brief claims that the lab used the identification criteria of TD2003IDCR including the 1% standard. The short answer to 2) is that they mostly used TD2003IDCR either including the 1% standard, or using a more relaxed standard.
[MORE]
1. Factual Background: The lab performs a GCMS on a reference material containing all 4 of the metabolites to be identified, here Cal Mix and blank urine, and also a GCMS on the Landis sample. Retention times, spectral and other data are measured for these GCs and compared with the corresponding RTs in the Landis samples. If they match an identification is found. Next the lab performs a GC-IRMS on the same Landis samples, the same blank urine, and on a Cal Mix that contains 3 out of the 4 metabolites to be measured. In the GC-IRMS the substances are burned up, so spectral data which aids in identification is lost. At this point the RTs may be compared and an identification can be made. The testimony and record are confusing as to this last step. Which RTs are being compared with which other RTs? Meier spoke of comparing the RTs of the GCMS against those of the GC-IRMS. Brenna, Ayotte, and Mongongu spoke of comparing the relative retention times (RRTs) as measured from an internal standard substance, and not comparing the RTs directly with the metabolites in the Cal Mix or blank urine. The majority decision says the RTs were compared directly to the reference substances and matched.
2. TD2003IDCR says among other things that one way to identify a substance is to match its RT with the RT of that same substance known to be in a reference material, or a urine sample “spiked” with that subtancel. It states that RTs must match within 1% although this limit can be relaxed, and it appears that this applies to RRTs also.
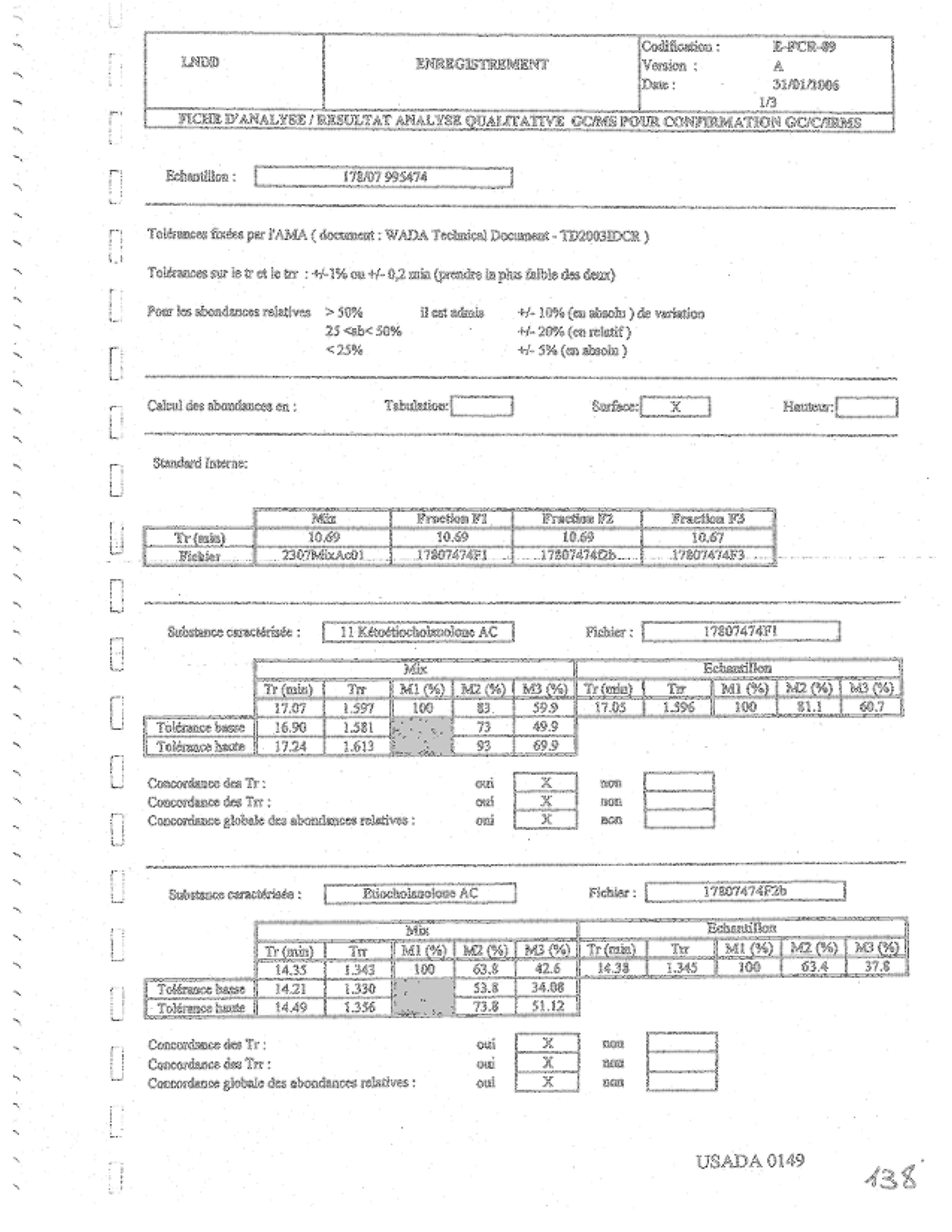
3. The USADA Pre-Hearing Brief stated (p. 23) that the lab used TD2003IDCR as their identification criteria, including the 1% standard, and that this applied to RRTs as well as RTs. When you look at the lab docs cited, (USADA 0149-51) it makes explicit that the lab was using this criteria, with the assumption that the GCMS matching of RTs and RRTs was sufficient to satisfy the identification requirements of TD2003IDCR and validate the identification in the CM-IRMS. The RTs matched. Since I couldn’t read the final briefs I wasn’t sure whether USADA argued this position after the hearing testimony.
USADA’s Pre-Hearing brief stated:
“Once the retention times and ion ratios were all entered in the form, the LNDD criteria for compound identification were applied. The LNDD criteria for compound identification are the WADA criteria defined in the WADA Technical Document entitled Identification Criteria For Qualitative Assays Incorporating Chromatography and Mass
Spectrometry, TD2003IDCR (Exhibit 12, page 2), as shown on page USADA 0149 (page section immediately below sample number):
ToldrancesJix~eps ar 1 'AMA (document : WADA Technical Document - TD2003IDCR)Meaning:
Toldrances sur le tr et le trr : +/- 1 % ou +/-0.2 min (prendre le plus faible des deux)
Pour les abondances relatives >50% il est admis +/-I 0% (en absolu) de variation 25 <>
Tolerance defined by WADA (document: WADA Technical Document TD2003IDCR)
Tolerance for the RT and RRT: +I- 1 % or +I-0.2 min (whichever is smaller)
Relative Abundance >50% : acceptable +/-I 0% variation (absolute)
25 to 50% +I-20% (relative)
<25%>
“The retention time or relative retention time of the compound in the sample must fall within the range defined by the retention time or relative retention time (respectively) of the reference compound, plus or minus the required value. The ion ratio for each ion in the compound in the sample must fall within the range defined by the corresponding ion ratio for the reference compound, plus or minus the required value. The evaluation of all data against criteria was all confirmed and documented on USADA 0149-0151 (Exhibit 24). Thus, LNDD identified the six compounds in Respondent's sample according to WADA TD2003IDCR.”
(USADA pre-hearing brief at 23-25)
USADA’s brief points to USADA 0149 as a summary of the identification data and criteria. USADA 0149 states in French that the document contains the GCMS qualitative analysis for the GC-IRMS confirmation, and that TD2003IDCR constitutes the identification criteria. This document indicates that the matching of the RTs and RRTs in GCMS against the reference material, Mix Cal Acetate, also constitutes the TD2003IDCR identification data for the GC-IRMS. That is, there is no direct matching of the GC-IRMS RTs or RRTs against the reference substances in the Cal Mix. If that is the proper interpretation, then one can argue that this identification procedure complies with TD2003IDCR when we relax the 1% requirement.
4. The majority opinion stated that the lab complied with the TD2003IDCR 1% standard for both the GCMS and GC-IRMS taken individually, but not across machines as Meier was arguing.
““Furthermore, as Dr. Meier-Augenstein attested, the RTs measured for the
GC/MS instrument and the GC/C/IRMS instrument separately are within the
1% criteria. There is no dispute on this point between the parties.”
I could confirm this for the GCMS, and now I think I have been able to confirm this for the GC-IRMS. USADA 0360 and 0362 show the GC-IRMS Mix Cal runs for the B sample and show RTs of: a) 870.6, b) 1241.8, c) 1316.7, d)1491.1. These RTs correspond to in the blank urine and Landis sample respectively: a) the 5A andro internal standard, b) the Etio in the F2 sample fraction, c) the 5B Andro metabolite in the F3 sample, d) the 11 keto in the F1 sample. So the Cal Mix contained a metabolite from each sample fraction, F1, F2, F3, but not the 5A Andro metabolite in F3.
Further it appears from USADA 351 that the RTs for all four of the metabolites in the sample match the corresponding RTs for the blank urine. It can be argued that the blank urine can be treated as a spiked urine sample and qualifies as a reference material for purposes of compliance with TD2003IDCR, although it does not appear that the lab treated it as such.
Similar documents exist for the IRMS on the A sample as well.
5. It appears that the lab has in fact matched the RTs of the samples with the RTs of at least 3 out of the 4 metabolites in the reference material or Mix Cal. The 5A Andro was not included in the Mix Cal so was not matched. The 5A andro is the metabolite that tested positive for exogenous testosterone. Nevertheless, there can be no doubt that the 5A andro was properly identified. The Mix Cal properly identified the 5B Andro in Landis’ F3 sample, and we know from the GCMS and the GC-IRMS urine blank that the 5A Andro directly follows the 5B by about 30 seconds. In the Landis F3 sample, the 5A Andro peak also directly follows (with no intervening peaks) the 5B by about 30 seconds.
6. So how reconcile all the testimony by Brenna, Ayotte, Mongongu, and even Meier about the use of RRTs measured from an internal standard. I don’t know. My guess is that the process described by Ayotte and Brenna is actually the way most labs do it. Even Meier didn’t say, look the proper way to do this is to compare the RTs of the GC-IRMS directly with the RTs of the Cal Mix. Instead he assumed the proper method was to compare the GCMS RTs with those of the GC-IRMS RTs. I really believe we’re missing something here because we are not scientists. None of the scientists either in the Landis case or on DP seem to have a problem with using RRTs in the GCMS and then matching those peaks in the GC-IRMS. It’s only anal lawyers. The other possibility is that the ID procedure in TD2003IDCR is not thought by scientists to be applicable to IRMS, or not understood well. Labs have developed their own procedures, with only lip service to the TD2003IDCR. This notion is supported by the fact that a future IS entitled “Reporting Guidance for Gas Chromatography/Combustion Isotope Mass Spectrometry Reporting Guidance” is being planned.
7. Conclusion: The retention times and relative retention times in the GC-IRMS matched those in the blank urine, which may be considered a spiked urine sample within the meaning of TD2003IDCR, and substantially matched those in the Mix Cal, which can be considered a reference material within the meaning of TD2003IDCR. Alternately, or in addition, the relative retention times of the GCMS matched those in the GC-IRMS in compliance with TD2003IDCR when we relax the 1% matching requirement. In all these cases, the identification criteria was TD2003IDCR itself.
Larry said...
M -
I appreciate the fact that you're carrying a heavy burden here, trying to deal with me and Mike S. at the same time, plus being about the only person on TBV willing to defend the FL decision. Like Mike S., I'm grateful for your presence here. Otherwise, Mike S. and I would be left "preaching to the choir", so to speak.
But I need you to take a more careful look at Procedural Order No. 2, and on the WADA rules that underline Procedural Order No. 2. This Order is not unique to the FL case; this order presents the "Catch-22" that every accused athlete must confront in a doping case. This order is not something that the FL team "agreed to" -- it is an order that was forced upon them.
I'll walk through the history that led to Procedural Order No. 2.
1. FL's document request to USADA is contained in a letter dated October 16, 2006 from Howard Jacobs. You can see this letter at http://trustbut.blogspot.com/2006/11/rejected-doc-request-correspondence.html. If you review this letter, you'll see that there was NOTHING that the FL team didn't ask for! The items that the Fl team asked for included the following:
- Any Standard Operating Procedure or SOP used by LNDD related to the processing of sample 995474 by GC-C-IRMS.
- All documents that evidence, reference or relate to the current IRMS criteria used by LNDD for determining an Adverse Analytical Finding.
- All documents that evidence, reference or relate to the identification of each of the peaks in the IRMS analysis of sample 995474.
- All mass spectral data necessary to identify all peaks within the MSD TIC analysis in connection with sample 995474.
- All data that has been used to identify the peaks in the IRMS analysis in connection with sample 995474, including any relevant isotope standards not provided within the laboratory documentation provided to date.
- All documents that evidence, reference or relate to the selection of metabolites used by LNDD for the carbon isotope ration test for testosterone using any GC-C-IRMS method.
- Please identify the precise time at which each peak on the MSD TIC scan appears.
I encourage you to read the Jacobs letter yourself. He asked for EVERYTHING, believe me. So we can both put aside the idea that somehow, the FL team did not ask to see the TD2003IDCR criteria.
2. USADA responded to the Jacobs request by letter dated November 3, 2006, as follows (see http://trustbut.blogspot.com/2006/11/no-documents-for-you-correspondence.html):
"After extensive review by us of your voluminous requests, I am writing to inform you that we will not be providing any documents or other information in response to your requests. As you should know, the rules applicable to this proceeding establish the set of documents that are provided by the laboratories when a sample tests positive. After studying your requests and those rules, every request you make appears to seek documents or information not called for by the rules."
In short: Jacobs asked for everything, and USADA said he'd get nothing. Nothing, that is, except for the documents in WADA's standard document package.
3. The arbitrators first got into the act on February 2, 2007, when they issued Procedural Order No. 1. In that order, the arbitrators deferred making any rulings on discovery matters:
"The Panel has also been advised that a disagreement has arisen between the parties concerning the discovery of documents. Counsels are to make further submissions on this matter after which the Panel will make its determination and any necessary orders."
4. We then reach Procedural Order No. 2, which I discussed in an earlier post. Your understanding of Procedural Order No. 2 seems to differ from mine, so let's look at it in depth. (http://www.usocpressbox.org/usoc/pressbox.nsf/6272c9a938d3a5cb8525711000564abd/9009e75c100f0679852572db006438fa/$FILE/Procedural%20Order%20No%202.pdf)
(a) Procedural Order No. 2 relies on WADA's Technical Document TD2003LDOC on Laboratory Documentation Packages. (See http://www.wada-ama.org/rtecontent/document/lab_docs_1_3.pdf) I won't quote from this TD, but will only point out that the TD does NOT require a lab to disclose any of the criteria it has developed under the ISL. The TD requires the lab to produce the documents we've all seen during the arbitration and on this site: collection control forms, internal chain of custody documentation, test results and the like.
(b) Under Section 7.1 of the ISL as well as the TD, the Laboratory is not required to provide any documentation not specifically included in the Laboratory Documentation Package. Section 7.1 of the ISL reads as follows:
"the Laboratory is not required to support an Adverse Analytical Finding by producing, either to the Testing Authority or in response to discovery requests related to the hearing, standard operating procedures, general quality management documents (e.g., ISO compliance documents) or any other documents not specifically required by Technical Document on Laboratory Documentation Packages. References in the International Standard for Laboratories to ISO requirements are for general quality control purposes only
and have no applicability to any adjudication of any specific Adverse Analytical Finding."
M, you stated that "SOP is not the same as 'identification criteria'. I believe they were entitled to discover this [identification criteria]." Sorry, but you're wrong on this point. It should be clear from the foregoing that the FL team was prohibited from discovering LNDD's documentation of its identification criteria.
(c) Procedural Order No. 2 added its own twist to the restrictions placed on the FL team by ISL 7.1 and TD2003LDOC. The WADA rules prohibit production of documents other than those listed in the TD -- the majority arbitrators went a step further and prohibited any testimony on the content of documents:
"a. A discovery response that there is no document will preclude the production of the document, along with any related testimony through a witness at the hearing.
b. A discovery response that there are documents that exist but they are not required to be produced because of the ISL or the other principles set out at paragraph 2 above will preclude the production of the document along with any related testimony through a witness at the hearing.
c. Any document used to determine the ISO certification or the WADA accreditation of the LNDD is not subject to discovery."
M, you stated that "They [the FL team] were not prevented from asking the witnesses about the identification criteria." Again M, I think you're wrong here. Procedural Order No. 2 states that the FL team was prohibited from seeing the written identification criteria and from seeking "related testimony" concerning the criteria. The phrase "related testimony" could not be much broader! How could they ask witnesses about the lab's identification criteria if they were barred from seeking testimony "related" to this identification criteria?
M, you stated that "in fact they [the FL team] did ask Mongongu and Brenna about this [the identification criteria]." M, I say this with all due respect: you're wrong on this point. Please revisit the cross-examination of Brenna in Brenna's first round of testimony -- the FL team never asked him about the LNDD's identification criteria. Why? Because they were prohibited from doing so under Procedural Order No. 2. Please revisit the cross-examination of Mongongu. Suh was able to ask Mongongu what steps she took, when she took those steps and how long it took to perform each step. He did not ask her anything about the criteria she used. why? Because he was prohibited from doing so.
Now ... you raise the point that, according to Procedural Order No. 2, the prohibitions on testimony described above were contained in "agreements reached covering the documentary discovery process." You infer from this quote that the FL team agreed to these restrictions. However, there's no indication in Procedural Order No. 2 that the FL team was a "party" to these agreements. The order simply says that agreements were reached. Perhaps the agreements were among the arbitrators. It's simply not clear from the Order.
In any event, I'm not sure why it would be relevant to our inquiry whether these restrictions were 100% forced on the FL team, or whether the FL team had any ability to shape these restrictions. The point is, the restrictions were there. They were quite real.
FL's team could not get written copies of the LNDD criteria.
FL's team could not ask questions about the content of the LNDD criteria.
FL's team was forced to do what I've been doing (and interestingly, what you've done in your latest post on the TD2003IDCR criteria): they were forced to infer the contents of the TD2003ICDR criteria: by asking the LNDD witnesses what steps they performed to identify the IRMS peaks, by asking the USADA experts how they would go about identifying IRMS peaks in this case, and by looking at the documentary package to see how LNDD appeared to identify the IRMS peaks. Under the circumstances, this was the only way open to them. And unless we're simply going to say that a lab's identification of IRMS peaks is presumed valid under all circumstances and regardless of the evidence, this is the only way open to us.
Larry said...
M -
Regarding your post claiming that LNDD actually and successfully used the TD2003IDCR standard ... wow. From my non-scientific background, I don't see how that can be. Dr. M-A and Dr. Brenna missed that? All the science types here and on DP missed that? Even OMJ would have missed that. I guess it's possible ...
I have not focused on how LNDD identified peaks in the GC/MS test, but I always assumed that they used RTs against a reference sample run contemporaneously on the GC/MS. I don't think there was any controversy about this, and I think this would explain the USADA brief on RTs and the statement in the majority opinion that the GC/MS RTs matched within 1%. In any event, I'm not trying to argue here that there was anything wrong with the GC/MS identification.
My understanding (and again, I'm no scientist) is that the LNDD did not identify the FL IRMS peaks by comparing them to a reference sample run contemporaneously with the FL sample. I'll put aside all of the testimony from Mongongu, Ayotte, et. al. to the effect that the IRMS peaks were identified by comparing them to the GC/MS peaks. You've raised this issue yourself in your point 6 -- I'll just point out the following:
- if LNDD was actually identifying IRMS peaks by comparing them to the peaks shown in a contemporaneous IRMS reference sample, then there would have been no need to complete a GC/MS test as part of the IRMS procedure.
- my understanding (based on the Mongongu testimony) is that LNDD does run a reference sample (I believe this is the mix cal acetate) through the IRMS, but this is for the purpose of calibrating the instrument (see testimopny p. 430, pdf p. 308). I suppose that the results of a calibration run, but that does not appear to be the purpose behind LNDD's use of the mix cal acetate.
- my understanding is that the results of the mix cal acetate run are shown in USADA exhibit 361, which you can find quickly at http://trustbut.blogspot.com/2007/09/retention-times-ii.html. According to TBV, the mix cal acetate run contained the 5aA internal reference and one of the three metabolites, 5bA. The mix cal run used by LNDD was missing the other two metabolites, 5aA and 5bP. Interestingly, if I'm reading the test results correctly, FL's sample was negative for 5bA. The positive finding, I think, was based on the metabolites not contained in the mix cal acetate.
I'll leave the rest to the science types here to figure out. My only additional comment is in reference to your comment about "anal lawyers." Aren't you GLAD we don't have any of THOSE kind of lawyers around here?
m said...
Correction to my last post.
The 5A Androstandial was found positive in the IRMS test, not the 5B Androstandial. My mistake. Can't keep them all straight.
6:30 PM
[BACK FROM MAIN BODY]
39 comments:
Comments here now.
TBV, can you post here M's comment from the old area, from 4:06 PM? That's probably the better place to start. My 5:35 PM post is a reaction to M's 4:06 PM post. It's his post that's the lead here.
OK, will do.
I assume we really meant M's 11:47; the 4:06 was Larry's.
Yes, thanks.
M, I think you're absolutely right that the LNDD should have identified the IRMS peaks for FL's S17 sample by comparing the peaks to contemporaneous peaks shown on an IRMS reference sample. That's the right way to do it. That's what TD2003IDCR seems to require. That's what TBV has been saying for at least a month now. And if you've discovered that this is actually what LNDD did in this case, and that they did it within a 1% margin, great! Then the issue of IRMS peak identification goes away, at least as far as I'm concerned.
However, I think there are some mistakes in your analysis - not surprising, as you're not a scientist. Neither am I, of course. But let me point out a couple of things:
1. In the process of trying to prepare a post on the meaning of the Brenna rebuttal testimony, I've been spending time carefully reading Dr. M-A's cross-examination.
In your point 4 in your 11:47 AM post on 10/31, you quoted the majority opinion to state that Dr. M-A attested that the RTs measured separately for the IRMS instrument were within the 1% criteria establised under TD2003IDCR. I think this is, indeed, what Dr. M-A said, but I think you may be misinterpreting what he meant.
(Note to all the science types here: please weigh in and correct anything I'm getting wrong.)
I think it's clear from Dr M-A's testimony that he was unable to identify any of the IRMS peaks used by LNDD as proof of doping: not any of the four metabolites that were used to show the presence of exogenous testosterone, not even the reference metabolite used as a RT baseline. I think that Dr. M-A testified only that the mix cal acetate runs performed during the IRMS testing had consistent RTs for each of the metabolites contained in the mix.
On this, I'll quote from Young's opening statement: "What mix cal IRMS is it's a solution of alkanes. There's four of them. And the reason that you run them is you run them three times in a row and see what kind of precision you get amongst the three times that you run it." M, THIS is where Dr. M-A testified that LNDD's RTs met the 1% standard: the metabolites in the mix cal acetate runs had RTs matching each other within the 1% standard. Dr. M-A did NOT testify that anything matched the metabolites in the FL samples within a 1% margin.
You can take a look at his testimony on this point and judge for yourself. It's at p. 1508 of the transcripts, pdf p. 1283. You need to read this testimony in context to get its meaning.
So ... I don't think you can draw on Dr. M-A's testimony to support you here.
2. You state in your paragraph 5 that the lab matched 3 out of the 4 metabolites. To do this, of course, the LNDD would have had to have at least 3 of the 4 metabolites in their mix cal acetate tests performed contemporaneously with the testing of FL's S17 sample. Moreover, we'd expect that the two metabolites used as the basis of the doping finding would have been included in the mix.
For ease of reference, let's set forth some of the science details. The 4 metabolites being measured by LNDD in their IRMS analysis are androsterone (andro), etiocholanolone (etio), 5-alpha-androstandiol (5aA) and 5-beta-androstandiol (5bA).
To make things a bit more confusing, FL's sample is divided into 3 fractions. If I'm following the science correctly, we can ignore the first fraction (F1). The second fraction (F2)contains the andro and etio, and the third fraction (F3) contains the 5aA and the 5bA. A reference standard compound, 5-alpha androstanol acetate (5-alpha AC) is added to all three fractions.
The main question here is, how many of these metabolites were contained in the mix cal acetate that M would like to use as the basis of his RT analysis?
We know from the cross-examination of Brenna during his first round of testimony that the mix cal acetate did not contain 5aA. (transcript p. 320, pdf p. 208). We also know from the cross-examination of Brenna in the second round of his testimony that the mix cal acetate lacked not only the 5aA, but also the andro. On this basis, Dr. Brenna testified that it would be impossible to calculate the RTs for the 5aA and the andro from the mix cal acetate (transcript pp, 1957-58, pdf p. 1690).
Finally, M, please note that LNDD's doping finding against FL was based on the results of its IRMS testing on 5aA and andro. FL's S17 results for etio and 5bA -- the two metabolites that LNDD could have identified by reference to the mix cal acetate RTs -- were within acceptable limits.
So, it appears to me that LNDD could not have identified the two key metabolites in this case by comparing FL's S17 sample to the mix cal acetate test results, using RTs or even RRTs
Like I said, I'm not a scientist, I may be missing something, and maybe you can reconcile the above with your theory that LNDD used RTs mostly within the 1% standard to identify IRMS compounds.
Larry--Duckstrap here. I've spent so much time reading your posts, I've not had time to comment--not that there has been a real need. Very interesting. I only wanted to chime in to say that I believe you have summarized the problems with the lack of the target metabolites in the IRMS mixed cal standards quite nicely. Further, since the IRMS has no identification capability, the need for chromatographic conditions consistent in every detail with the GCMS (i.e. within the 1% RRT) is required to assure that what is identified in GCMS is the same as what is measured in the IRMS. Furthermore this condition is needed to assure that no other peaks have moved around such that they are now interfering with the putative CIR measurement.
Kevin/Duckstrap,
Ok, I'll ask you this again. There does not appear to be any other possible mapping for those 4 central peaks in the GC-IRMS of the F3 other than the 5A and 5B. I believe you have at one time admitted this. Yes, in the absence of the full mass spectra, there is some very remote possibility that they might contain some other substance that elutes at exactly the same time as the 5A and 5B and is hidden in those peaks, but there is almost no doubt that it contains the 5A and 5B.
Perhaps you could also explain why the blank urine cannot serve as a "spiked" urine sample for purposes of comparing the retention times of the 4 metabolites.
Larry,
I've sent to TBV an updated post about the identification which has more detail and some changes, and explains better my analysis. I'd rather wait until that is posted before responding.
The Mix Cal Irms is different from the Mix Cal Acetate and is used for a different purpose.
Kevin-Duckstrap -
Wow. If you barely have time to READ my posts ... then I'm spending way, way too much time writing them! Worse, if I'm beginning to understand the science, I may lose the right to ask dumb questions.
Notwithstanding, I've got another series of dumb science questions, and I'll ask most of them by stating my understanding of the science and asking if you (and the other science types here) can confirm or deny.
When we look at a GC/MS graph, the height of the peaks in the graph (or more accurately, the area described by the peak) represents the amount of a particular metabolite we're trying to measure? And it's possible by looking at the two-dimensional shape of the peak and its RT to identify the metabolite?
In contrast, when we look at the peaks on the IRMS graph, the height of the peaks (and NOT the area described by the peaks, as the peaks appear to be one-dimensional to me) represent not the amount of a metabolite, but the C13/C12 ratio for the metabolite (the higher the peak, the more C13)? And because the peaks are one-dimensional, the peak contains no identifying information other than its RT, which in all cases has to be matched up to a known reference if the peak is to be identified?
And just to make this as clear as I can: there could be a tiny amount of a particular metabolite present in a sample that would create a large IRMS peak, and a huge amount of a particular metabolite in a sample that would create a tiny IRMS peak (or in theory at least, no IRMS peak whatseover!)?
Out of curiousity: is it possible that any of the metabolites of interest to us in this case might have had no C13 content, or so little C13 content that it would not create a noticeable IRMS peak?
My guess is that, if we're looking at molecules in the human body containing carbon, a certain percentage of these molecules in ANY human body will contain C13 and show up in an obvious manner on an IRMS chart ... but so long as I'm asking questions, I thought I'd ask this question too.
Back to the main point: if there's no relationship between the height of a GC/MS peak and the height of an IRMS peak ... then what the heck was Brenna talking about when he advocated "pattern matching"? What pattern can you match? Under the best of circumstances, there's not going to be a pattern along the y-axis, just a pattern on the x-axis. And the x-axis are RTs ... and Brenna said you could not match RTs, so he's saying that there's no pattern on the x-axis either. Is there another dimension I'm missing that Brenna uses to find his pattern?
(the last question was intended to be rhetorical, not dumb, but it might classify on both counts!)
M, OK, then I'll wait to see your next post. You're right that I may have mixed up the various reference samples, but if I did, it also looks like Dr. Brenna did the same in his testimony.
OK, quick correction to my last post: I made the mistake of saying that the height of a peak on an IRMS chart indicates the C13/C12 ratio for the peak. Sorry, that's clearly wrong. I'll let my questions stand from my prior post, as they may help explain things to those of us who struggle with the science. But clearly, I could have asked a much better set of questions.
I am not sure if this is the appropriate place to ask these questions, but here goes. Obviously the science in the appeal to the CAS will be complicated, at least as complicated as the case presented to the USADA arbiotration panel in May. Who may I ask, will be "explaining" the scientific issues to the CAS panel? Dr. Botre? I hope not! And any idea of when/if we will know who is selected for the panel? Thanks, you may now go back to your arcane discussion, the fool has left the room.
str
The fool has left the room? I thought I was still here.
syi
Duckstrap here. I'll try to give my take Larry's comments:
Larry –“When we look at a GC/MS graph, the height of the peaks in the graph (or more accurately, the area described by the peak) represents the amount of a particular metabolite we're trying to measure? And it's possible by looking at the two-dimensional shape of the peak and its RT to identify the metabolite?”
In the GC/MS, the identfication criteria is supposed to be the mass spectrum of the particular peak, or more properly, the abundance ratio of the diagnostic ions. These are the mass spectra that LNDD never supplied. They would show whether the target peaks in the GC/MS contain only the substance of interest, but nothing else. So we should be getting two things from the GC/MS: 1) a positive identification of a target peak, and 2) its RT (whether relative or absolute). The retention time is what allows the peak that was positively identified by its mass spectrum to be linked to a putatively corresponding peak in the IRMS.
Ideally, for this link to be maintained, the chromatographic conditions should be essentially identical in the GC/MS and IRMS instruments. That way you could be assured that the separations were the same, and that the peak identified by GC/MS had the same characteristics in the IRMS before it was all burned to CO2 and water. If the chromatographic conditions are not the same, then there needs to be an alternative method for evaluating what is what in the IRMS, i.e. RT or RRT. LNDD’s chromatography was different between the two instruments—thus there needed to be some systematic way of evaluating the identification.
In the GC/MS, it is the amount of charge in a particular peak that gives it its height and area. Many substances have a single electron knocked off by the ionization source, and thus have a single positive charge. Others end up with 2 positive charges. A substance that has a mass/charge ratio of say 44/1 will have half the peak height for a given molar amount as something that has a mass/charge ratio of 44/2 = 22. Thus for GC/MS peak height alone is not necessarily an indicator of the amount or identity of what is in the peak.
Larry – “In contrast, when we look at the peaks on the IRMS graph, the height of the peaks (and NOT the area described by the peaks, as the peaks appear to be one-dimensional to me) represent not the amount of a metabolite, but the C13/C12 ratio for the metabolite (the higher the peak, the more C13)? And because the peaks are one-dimensional, the peak contains no identifying information other than its RT, which in all cases has to be matched up to a known reference if the peak is to be identified?”
I believe that what is displayed in the IRMS chromatograms in the lab package is the abundance of m/z (charge to mass ratio) = 44 versus elution time, corresponding to the abundance of C12 in the GC eluate. The C13 (m/z=45) trace precedes C12 by about 150 ms, due to something called the “chromatographic isotope effect” and is not shown in the LDP. A third trace that is also not shown is the ratio of m/z = 44 to m/z = 45 vs. time. If you sum up the area under the C12 peak and the area under the C13 and take the ratio, that gives the carbon isotope ratio or CIR. Obviously, this ratio will be sensitive to how the instrument background is treated, and how the integration (summing up of area) is treated. LNDD used a certain amount of operator judgment in these areas. However, the identification of these peaks rests on their retention times compared to the GC/MS in some form or fashion. The peak heights are going to correspond to how much carbon comes from a given substance. Thus something with 22 carbons will have twice the peak height for the same molar amount as a substance with 11 carbons in the IRMS. So again, the peak height all by itself does not necessarily carry with it identifying information.
Larry - ”And just to make this as clear as I can: there could be a tiny amount of a particular metabolite present in a sample that would create a large IRMS peak, and a huge amount of a particular metabolite in a sample that would create a tiny IRMS peak (or in theory at least, no IRMS peak whatseover!)?”
A large molecule with many carbons per molecule and present in small amounts would be burned to produce a relatively large amount of CO2 with a correspondingly large IRMS peak. Conversely, a substance with no carbon at all could pass through undetected since it would produce no CO2 upon combustion.
Larry – “Out of curiousity: is it possible that any of the metabolites of interest to us in this case might have had no C13 content, or so little C13 content that it would not create a noticeable IRMS peak?
My guess is that, if we're looking at molecules in the human body containing carbon, a certain percentage of these molecules in ANY human body will contain C13 and show up in an obvious manner on an IRMS chart ... but so long as I'm asking questions, I thought I'd ask this question too. “
Now you’re on to Duckstrap’s favorite speculation. Part of the preparation for IRMS involves substitution of certain parts of the metabolites containing oxygen and hydrogen (hydroxyl groups) with groups containing carbon and oxygen (acetate groups). The acetate that was used to perform this derivatization contains almost no C13 (had a CIR of –53, per e.g. USADA 0351). The reason for this is not clear to me, though I am sure there is a good one. However, if something with more hydroxyl groups than the testosterone metabolites (which have 2 apiece) were treated with this derivatization reagent, then it would have a substantially more negative CIR than the testosterone metabolites. Now if one of these substances coeluted completely or partially with the testosterone metabolites, then the measured CIR would be badly biased in a more negative direction.
Larry – “Back to the main point: if there's no relationship between the height of a GC/MS peak and the height of an IRMS peak ... then what the heck was Brenna talking about when he advocated "pattern matching"? What pattern can you match? Under the best of circumstances, there's not going to be a pattern along the y-axis, just a pattern on the x-axis. And the x-axis are RTs ... and Brenna said you could not match RTs, so he's saying that there's no pattern on the x-axis either. Is there another dimension I'm missing that Brenna uses to find his pattern?”
That is a pretty darn good question, and I am responding in a rhetorical fashion, though it clearly has more than rhetorical significance.
To review: We have incomplete specific identification in the GC/MS step since the complete mass spectra were apparently never examined, were not provided in the LDP, and indeed have since been destroyed. We have chromatographic conditions in the IRMS that are not consistent with the system that was used to identify the metabolites. Therefore, there is no absolute retention time or relative retention time correspondence between the systems. The calibration mixture that was used to “identify” peaks in the IRMS did not contain the substance (5aA) that was found to be indicitive of doping. Finally, the peak height “pattern” used to identify the target peaks might be, but need not be, consistent between the instruments.
Kevin/Duck -
No pun intended by the way I've addressed this post, but here comes more stuff from me.
I've read a lot of discussion here about the missing mass spectra, now maybe I'm finally beginning to catch on. If anyone wants to see what the mass spectra of testosterone looks like, I have a link to it: Testosterone mass spectra. For the record, this mass spectra graph is taken from the article at OSHA Drug Discussion.
Interesting, Kevin. Now I can see how mass spectroscopy can actually IDENTIFY something. I have some follow-up questions:
1. When we're looking at the mass spectra for testosterone, presumably this is a spectra for all testosterone metabolites combined, and not for any single metabolite?
2. Is there an ISL standard governing mass spectra data? In other words, from the standpoint of the ISL, how would you determine that the mass spectra data actually shows that a particular substance is testosterone? Presumably, you'd compare the test data to a reference spectra. But does the ISL tell you how to make this comparison? Do you have to "eyeball" the test data against the reference spectra?
3. If my understanding in (1) above is correct, can you use the mass spectra to identify a given PEAK on a GC/MS graph? If so, how would you do it? Does TD2003IDCR address how this would be done?
Larry (and M, if you are still listening),
There is a reasonable thumbnail description of GC/MS and IRMS in Thomas Fine's Wiki at http://landiscase.wikispaces.com/GC+IRMS (sorry, can't figure out the links thing). The problems with coelution of different substances in chromatographic separations are well known. This is the essential problem with using only retention time to identify substances in a chromatogram and is the driving reason for coupling a mass spectrometer with chromatography (gas or liquid). Essentially, the MS in GC/MS is what identifies each of the peaks in the chromatogram, and it does so by looking at the mass spectrum (or parts of the mass spectrum) of each of the peaks. TD2003IDCR talks about this at some length. If both the retention time and the mass spectrum of a target substance match a standard or reference then you can be pretty sure that you have the right stuff (assuming, of course, that you have done your work correctly). LNDD was sloppy. First, they threw away the information in the complete mass spectrum that would have ruled out interferences in their analytical GC/MS. Then they changed the chromatographic conditions between GC/MS and IRMS, making the retention time consistency between the methods (highly) questionable. Then they used the "eyeball squint method" to do integration and background correction in the IRMS. Possibly an ok way to waste time in a PhD project, but I wouldn't want someone putting my livelihood on ice this way.
Kevin -
Thanks. I'll read your post about a dozen times, the way I usually do to realize that it's actually answered my questions. I'll also look at the cite you've given me.
I'm disturbed that you can master the science of mass spectroscopy but not how to do a simple HTML markup! Oh, well. Nice to know that I know something you don't know. May never happen again. ;^)
M -
Thanks for updating your post, where you argue that LNDD used the 1% standard under TD2003IDCR to identify the IRMS peaks that served as the basis for the doping finding in the FL case.
I posted some objections to your post this morning at 12:30 AM, and you stated that you'd wait to address these objections until after your post was updated. I don't think your update addresses the points I made about the correct interpretation of Dr. M-A's testimony. However, since you don't rely much on Dr. M-A's testimony to make your case, there's not a pressing need for you to respond to me on this point.
The major point we need to clear up concerns the number and the identity of the metabolites present in the reference samples run in the IRMS contemporaneously with the three FL fractions for S17. To reiterate from my 12:30 post: there are four metabolites of interest in the IRMS test used to determine the presence of exogenous testosterone in an athlete's sample: androsterone (andro), etiocholanolone (etio), 5-alpha-androstandiol (5aA) and 5-beta-androstandiol (5bA). FL's doping finding for S17 was based on 5aA (and, according to some of the arguments made by USADA, on the andro as well).
In your supplemented post, you claim that LNDD performed a GC-IRMS "on a Cal Mix that contains 3 out of the 4 metabolites to be measured." I argued at 12:30 AM that the mix cal acetate contained TWO of these metabolites, etio and 5bA, but did not contain 5aA and Andro (the two metabolites we need to identify to support the doping finding). You responded briefly at 7:01 AM that "the Mix Cal Irms is different from the Mix Cal Acetate and is used for a different purpose."
M, one or both of us may be mixing up terms, but I think we actually agree on what was in the mix that you're trying to use to identify the FL S17 IRMS peaks. We just disagree on how many of the 4 critical metabolites were contained in the mix. Since we don't agree on what to call this mix, let's call this the "reference mix" for the moment. You and I agree that the reference mix contained etio and 5bA, and we also agree that the reference mix did NOT contain 5aA (you've so stated in paragraphs 4 and 5 of your supplemented post).
The only question is, did the reference mix contain andro? I've argued that it did not, and offered proof from Dr. Brenna's testimony and from information posted here by TBV. I don't see anywhere in your supplemented post where you claim that the reference mix DID contain andro. In your paragraph 4, you claim that the reference mix contained the following metabolites (I've identified each metabolite in brackets, using the naming convention I've specified above : a) the 5A andro internal standard [this is 5aA, but since it's used as an internal standard, it can only be used to identify the internal standard added to FL's S17 sample, as you've implicitly acknowledged], b) the Etio in the F2 sample fraction [etio], c) the 5B Andro metabolite in the F3 sample [5bA], and d) the 11 keto in the F1 sample. I think you're right that the reference mix did contain these substances.
I did not identify the 11 keto with a bracketed reference, because I have not discussed 11 keto before. 11 keto (full name: 11-Keto-etiocholanolone) is NOT one of the four metabolites we've discussed up until now. 11 keto is one of two OTHER metabolites (the second is 5-beta-pregnandiol, or pdiol, which is also in the reference mix) used as an endogenous reference compound to compute the delta/delta values required to prove the doping violation. The identification of these endogenous reference compounds is not at issue in this case.
So, I'll reiterate: the reference mix you've referred to in your supplemented post contained only TWO of the four metabolites used to determine the presence of exogenous testosterone in FL's S17 sample. The reference mix did NOT contain 5aA or andro, which were the two metabolites used to convict FL. So you can't use the reference mix to identify the 5aA and andro in FL's S17 sample, at least not by any method described in TD2003IDCR.
There is much, much more that needs to be analyzed and discussed in your supplemented post. Hopefully, others will help me with this job! I think that the next matter to consider is your argument that the blank urine samples can be used as a reference to identify the IRMS peaks in FL's S17 samples. I'm not ready to tackle this job just yet!
Larry,
I said that the Mix Cal Acetate showed the RTs for the internal standard 5A Androstanol, and 3 out of the 4 metabolites, 5B androstandial, etio, 11-ketio. The only metabolite that was missing was the 5A Androstandiol.
This is correct.
1. Meier was dead dead wrong when he said the 5A internal standard was not in the Mix Cal. He must have had a memory lapse. Brenna explains this at transcipt 1657-1658 for the A sample. The Mix Cal's show that 5A eluted at 866 in the A sample and 870 in the B sample. This is evident from the exhibits, e.g. USADA 360 and 362. The 5A internal standard eluted at the same times in Landis' samples and the blank urine.
The cross-exam testimony that you reference at p. 208 is Brenna conceding that the 5A androstandial metabolite was not in the Mix Cal. He was not talking about the 5A internal standard.
BTW you notice that Duckstrap hasn't addressed my question about the peak identification
He poses general potential problems,but he doesn't show whether any of these problems likely occurred in this specific case. For example, he cannot show that the 5A or 5B peaks could have switched in this case. Remember we know that the 5B peak did not switch because it has the same retention time as in the Cal Mix.
Larry,
What you are calling the "andro", I'm calling the 11-ketio. So we're on the same page.
Larry and Kevin,
The potential for a peak to switch positions supposedly stems from the small variation in GC-IRMS ramp conditions from that in the GCMS.
The easy way to test this is to do a GCMS with the changed ramp conditions and see if the peaks switch. I'm sure they won't switch. Why? Because what we got in the IRMS chromatographs was a stretching out of the peaks. In any case this can be proved, so why doesn't Landis do it if its supposedly such an important possibility. Perhaps because he would rather argue the uncertainty.
And note that including both the 5A and 5B in the Cal Mix reference would not solve this problem. Why? Because if they switched in Landis' sample, they also would have switched in the Cal Mix and you couldn't tell which was which or whether they had switched at all. By including only 1 metabolite in the Cal Mix, you at least know that it's peak is properly identified and know its retention time.
Larry,
"I don't think your update addresses the points I made about the correct interpretation of Dr. M-A's testimony."
I said the majority opinion said Meier admitted that the RTs for the IRMS met the 1% standard.
I think Meier's testimony bears this out.
"Q. And that tells you that your instrument, when it identifies the retention time of the internal standard, it's identifying the retention times of the other metabolites or
analytes in question.
A. On that instrument, yes.
Q. All right. And then, when you looked at the retention times and relative retention times on the IRMS instrument, they were all very consistent too, weren't they?
A. Yes.
Q. And so the comparison that you were making was not within a particular instrument?
A. No, that's incorrect. This comparison would have been the instruments -- within the instruments as well as between the instruments.
Q. And -- well, within the instruments, there was great consistency within 1 percent?
A. Yeah."
YEAH!
M -
Sorry, I don't think we're on the same page yet. Andro is not 11-ketio. They're different substances.
Andro is a testosterone metabolite used to test for the presence of exogenous testosterone. When you do the calculation delta-1 minus delta-2 to find a difference in delta values (a -3.8 per mil difference being proof of a doping violation), Andro is one of the 4 delta-1 substances. It's the identification of those 4 substances that we've been talking about here.
11-ketio is a delta-2 substance used in the computation to find the delta-delta values for both etio and andro. These delta-2 substances are endogenous reference compounds. The identification of 11-ketio is not an issue in the FL case, at least not to my knowledge.
To prove the doping charge, USADA had to identify the 5aA and perhaps also the Andro. That's the stuff that was not in the reference mix. So they could not identify these two substances by comparing the 5aA and Andro in the FL samples to the peaks for the reference mix, at least not by any method explicitly recognized by TD2003IDCR.
I apologize if all I've done here is to state the obvious and if I've completely missed your point. I know that I have not responded to everything you've posted tonight, but we've GOT to get on the same page regarding the reference mix.
Larry,
I see your point now and agree that the Andro is one of the 4 metabolites and was not in the Cal Mix Acetate. So you are correct, only 2 out of the 4 metabolites were contained in the Cal Mix Acetate.
So what we had in the Cal Mix Acetate was the 5A internal standard, which was in each sample fraction, and then 1 other substance in each sample fraction.
BTW, while reading the majority decision to understand your points, I discovered that the Landis folks were treating the Mix Cal Acetate as a positive control or quality control wrt to the IRMS, not a reference sample for RTs.
On the other hand, the majority noted that the 5A internal control was added to the Mix Cal for purposes of calculating RRTs, but doesn't say what these RRTs are to be compared with. Presumably the RRTs in the GCMS.
So its possible that neither side thinks the Mix Cal is used as a reference material for direct comparison of RTs in the IRMS procedure.
I'm beginning again to doubt whether direct comparison of IRMS RTs to a reference material is how either side viewed the identification process as specified by TD2003IDCR.
Not that we aren't all confused from time to time, but...
The stuff in the IRMS mix cal is the 5aAC internal standard, the 11k-etio, the Andro, and the 5bA.
The targets in the F3 are the 5aAC, the 5bA, the 5aA, and the pdiol, of which ostensibly meaningful CIRs are taken of the 5bA, 5aA and pdiol. The pdiol is common to both the reported 5bA and 5aA delta-deltas, so it its value is off, so are the reported values of the 5xA's.
TBV
Larry,
Does TD2003IDCR require that the sample retention times match those of the reference material in the GC-IRMS? Answer: Probably Not.
TD2003IDCR specifies that the retention times of a sample must match the retention times of a reference material within some percentage standard. The question in this case is whether this refers to the reference material retention times in the GCMS or the GC-IRMS.
TBV and you have taken the position that the lab must directly compare the GC-IRMS retention times (RTs) of the metabolites in Landis' samples directly to the GC-IRMS retention times of a reference material, here the Cal Mix Acetate, in order to satisfy the identification criteria, stated in TD2003IDCR.
I've taken the position that this appears to be a possible method, but that TD2003IDCR also permits one to compare the GC-IRMS retention times and relative retention times of Landis' samples to the GCMS RTs and RRTs of the reference material, and that in that case one can relax the 1% standard. A third position is that one just needs to verify that the GCMS retention times of the sample match those of the GCMS reference material within the 1% standard, because the GC-IRMS uses the exact same sample. This appears to be what the lab documents show.
I now believe that latter two options are permitted by TD2003IDCR. I do not believe the standard requires the lab to match the RTs or RRTs of the reference material in GC-IRMS with those of the sample.
As I noted in my main presentation above, USADA seems to take this position in it’s pre-hearing brief. The lab docs (USADA 0149-0151) indicate this is the method the lab used to satisfy TD2003IDCR. Actually they show that the lab compared the GCMS sample RTs with the GCMS Cal Mix RTs and these matched within the 1%. I think the assumption here is that since the exact same sample was run through the GC-IRMS there was no need to compare the IRMS RTs with the GCMS RTs. However, Brenna did do so and found the “pattern match” but said the 1% tolerance didn’t apply.
Nearly all of the testimony was consistent with the comparison of the sample RTs with those of the reference material GCMS RTs.. Brenna, Mongongu and Ayotte and Meier all spoke of calculating RTs and RRTs from the internal standard and presumably comparing those RRTs with some reference which could only be the GCMS. The only exception was Meier's concession on cross that the IRMS RTs were "consistent" "within the 1% standard", and the majority opinion's characterization of this testimony that implied that there was a direct comparison of the sample RTs with the Cal Mix RTs in the GC-IRMS. As we have seen this occurred but with only 2 out of the 4 metabolites. While with the GCMS all 4 metabolites were included in the Cal Mix.
We've now seen that the Landis side viewed the Cal Mix used in the GC-IRMS as "positive control", and not as a reference material.
TBV once asked why didn't the lab include all 4 of the metabolites in the Cal Mix for the GC-IRMS, when it had done that for the GCMS. I think I have an answer now, although it’s a little complicated.
We've observed that the RRTs in the Landis GC-IRMS are off by about 4-6% from those in the Cal Mix reference material in the GCMS, probably because the ramp conditions were not exactly the same. Remember Meier said that its difficult to get the ramp conditions exactly the same. The Cal Mix for the GC-IRMS contains 2 of the 4 metabolites and 2 other substances. We’ve observed that the RTs for all of the substances in this Cal Mix match those in the GC-IRMS for Landis’ sample within the 1%. Now let's say the Cal Mix for the GC-IRMS contained all 4 of the metabolites. Well they would most certainly all have matched within the 1% standard also, but they still would have been off by about 4 to 6% from those of the GCMS.
How do we know that we can use the GC-IRMS Cal Mix reference sample as an identification reference? How do we know that the 5B in the Cal Mix really was the 5B that we had identified in the GCMS and not the 5A which was right next to it? Only by comparing it with the 5A in the GCMS. If the peaks in the Cal Mix were very far apart we could be confident that they corresponded to the same peak in the GCMS even though the RTs didn't match. So we would have to compare the peaks in the GC-IRMS with those in the GCMS anyway. We would still be using structural information to help us, i.e. the sequence of the peaks, how far apart they were, and the fact that we knew that the Cal Mix must contain the 4 metabolites. While there are more substances in the Landis sample than in the Cal Mix, we also know exactly which metabolites each sample contains because we had fractionated the sample.
Duckstrap could still argue that the peaks had switched in the IRMS Cal Mix, so that what appeared to be the 5A was really the 5B. But putting all 4 of the metabolites into the Cal Mix doesn't absolutely solve that uncertainty problem. In fact as I have noted, it's better to only have 1 of the metabolites in a sample in the IRMS Cal Mix, because then there is no doubt about the identity of that peak.
So the fact that the you’ve matched the RTs of the IRMS reference material with those of the sample within the 1% standard adds little additional information over matching the sample with the GCMS reference material. You still have to compare the RTs in the IRMS reference sample with those in the GCMS. It just adds another layer of confirmation and consistency.
So my position is:
1) The lab used TD2003IDCR as its identification criteria. This criteria was adequate.
2) Under TD2003IDCR the lab may match the GCMS RTs/RRTs of a reference material with those of the GC-IRMS Landis sample, but these do not have to match within the 1% tolerance because different GC machines are being used.
3) Under TD2003IDCR the lab may match the GCMS RTs/RRTs of a reference material with those of the GCMS Landis sample and these must match within 1%, so long as the GC conditions are substantially the same. This last argument needs some fleshing out.
4) TD2003IDCR does not require the lab to match the GC-IRMS RTs/RRTs of a reference material with those of a sample within the 1% standard or any standard.
M -
Good! We ARE on the same page.
Two points about Dr. M-A's testimony: (1) despite the translation, I suspect that the good Doktor was really saying "ja" and not "yeah". This doesn't change the substance of his testimony, but it makes the testimony sound more adult. (2) When Dr. M-A said that there was 1% consistency within the instruments, I think he meant that the mix cal acetate runs showed consistent results and not that any mix cal acetate result compared within 1% limits to FL's S17 samples. If this is an important point to you, I'll explore this further in another post; otherwise we should probably move on to other points.
On your point about Duckstrap not having answered your question ... my experience with Duckstrap (my good friend and ally!) is that he always answers my questions but he doesn't always answer them in the form I was expecting. I won't speak for you, but I ask Duckstrap science questions from my law perspective that he answers from a science perspective. Or ... maybe the better way to put this is: Duckworth takes the questions that I ask based on a relatively poor understanding of the science, rephrases them in his mind to make them better questions, and then answers the improved version of the question!
All of this is to say, in a nice way from one non-scientist to another, that I think Duckworth HAS answered your question. If you like, I can try to explain how he's done this.
I think I understand when you say that you're starting to doubt whether either side viewed the identification process as being the process specified in TD2003IDCR. As one lawyer to another: we're being asked to make sense of information that is a confused mess. As I've taken on the role here as defense counsel, I'll blame the LNDD and USADA for this mess; given the role you've taken on here, you'll probably blame the FL team for this. But in private conference, we can do what opposing lawyers do in private: that is, complain about our clients!
I'll try to post something more substantive later in the day.
For the record, my 9:57 AM post was composed and posted before I saw M's 9:46 AM post. So, please don't read anything I said at 9:57 as a response to what M posted at 9:46.
TBV,
Do I take it that I spent one hour going back through the exibits and confused myself again!
You mean I was right the first time and the Andro was in the Mix Cal!
I give up.
Oops, I'm wrong. No andro. The simple answer is on USADA 354 where the cal mix is measured for the CIR's of the 5aAC, Etio, 5bA, and the 11-k.
TBV
And addendum to my 9:46 am post.
What is the purpose of the IRMS Mix Cal acetate, if it's not a reference material to match RTs for identification. Unlike the GCMS Mix Cal it cannot be used to identify a substance since there is no associated mass spectra.
I think it is a quality control aid, in part to show that the RT matchs with the GCMS reference material are accurate. It provides a couple of accurate IRMS RT anchor points in each sample from which to check the location of the IRMS peaks and their sequence. This aids in linking them with their GCMS counterparts which will have a slightly different RT and RRT for the reasons given by Brenna.
M -
Responding to your 12:17 pm post, yes the mix cal acetate is used as a quality control, or what Dr. Brenna called a "positive control".
The purpose of these controls is to verify that the IRMS unit is working properly. You're looking to see that the IRMS machine delivers the same (or close to the same) numbers for the metabolites present in each control.
According to USADA's opening statement in the arbitration transcript, LNDD actually ran three different IRMS controls: a mix cal IRMS, the mix cal acetate we've spent so much time figuring out, and the blank urine.
The sequence of when each control was run appears to have been a matter of some confusion in the arbitration testimony. From what I can see, the IRMS process starts with three mix cal IRMS injections. Mix cal IRMS is a solution of 4 alkanes. My online chemistry dictionary says that an alkane is a hydrocarbon containing only single carbon-carbon bonds, such as C2H6. I can't find any place in the testimony where these alkanes are identified, but it seems obvious that none of these alkanes could resemble the metabolites we need to identify to prove a doping violation. You could not use the mix cal IRMS to identify anything of interest in the FL case.
Once the mix cal IRMS test was completed, LNDD then ran a mix cal acetate, then blank urine, then FL's F1 fraction, then blank urine, then F2, then blank urine, then F3, then a final mix cal acetate. Or maybe the fractions were run before the blank urine controls, I'm not 100% sure.
The important point to note here is that the control injections are NOT run to identify peaks in the FL samples. The purpose of the controls is, initially, to make sure that the machine is operating properly, and then during the test, to make sure that the machine is maintaining its precision and accuracy. Moreover, the peaks for each kind of control are compared only to other runs of the same control. So, for purposes of the control you'd compare mix cal IRMS peaks to other mix cal IRMS peaks, mix cal acetate peaks to other mix cal acetate peaks, and blank urine peaks to other blank urine peaks. Within each type of control, the peaks should be consistent. (This is what I think Dr. M-A meant when he said there was great consistency within the instruments.)
Now as TBV has maintained, the LNDD MIGHT have used the mix cal acetate runs not only as a control but also as a means to identify the FL IRMS peaks. But they did not do so. If they had intended to do so, then they would have included the 4 critical metabolites in the mix cal acetate.
The remaining question is whether the urine blanks could be used to identify the FL IRMS peaks. I know you've maintained that this might be possible. Urine blanks are "live urine" (in the words of USADA in its opening statement), not a chemical solution. The blank urine is intended to resemble the urine matrix of the athletes. The blank urine is a useful control because the lab can compare the blank urine runs to runs made previously by the lab, and the lab can determine if the current runs are consistent with the blank urine runs that the lab has obtained historically. So if the chromatography for a current blank urine run is not as good as the blank urine chromatography that the lab has obtained historically, that's probably a good indication that there's something wrong with the IRMS machine.
Can you identify the IRMS peaks from an athlete's sample by reference to the IRMS peaks in a urine blank control? One clear answer: no such identification could satisfy the criteria specified in TD2003IDCR. TD2003 IDCR requires RTs to be measured against "spiked urine samples", "Reference Collection" samples or "Reference Materials" (the latter two terms are defined in the ISL). The blank urine is "live" and not spiked, and it's not designed to be used as a reference to identify substances. Blank urine is designed to look like real urine. The reason LNDD uses blank urine as a control is because its other control mixes are artificial mixes DESIGNED to serve as reference materials, and LNDD appears to have wanted to use a more real-world control substance that was NOT so designed.
So ... hopefully I've cleared up what these "quality controls" are intended to accomplish, and I've thrown some doubt on whether any of these controls (in particular, the blank urine controls) could be used to identify an athlete's MS peaks. I apologize for not providing page and document cites -- I can do so if anyone needs to see them.
I want to correct one statement I made in my 2:59 post. I said that blank urine is not spiked. That's not exactly true. LNDD DOES add the 5aA-AC internal standard to the blank urine. I think they do this to allow the RTs for the blank urine peaks on a given IRMS chart to be accurately compared to the RTs for blank urine peaks in other IRMS charts. On this point, see the Mongongu testimony, p. 653 of the transcript, pdf p. 509.
This does not detract from the point I'm trying to make: when LNDD2003IDCR calls for the use of a "spiked" sample, it means that the sample should be spiked to allow for identification of the metabolites in an athlete's urine sample.
Larry,
I agree with the gist of what your 2:59pm post. I recall reading much of that before, but it's all starting to blend together now. I'm going to take a break, despite and maybe because of the new document dump.
I tend to agree that the blank urine shouldn't be used as a GC-IRMS RT reference material (spiked urine sample) for the same reason I don't think that the Cal Mix Acetate even if it had contained all the metabolites should or could be used as an IRMS reference sample. Since there are no mass spectra, if you include all the metabolites, some of them are so close together that you can't be sure if you've identified them correctly by retention times alone in your Cal Mix.
It's possible to use the Cal Mix Acetate to fix the RT of what the arbs called the chromatographic standard, 5A androstanol, and from this one can calculate the RRTs of the metabolites for comparison with those in the GCMS. Its also possible to use the Cal Mix to fix the RT of one of the metabolites if it's RT is sufficiently distant from that of any other metabolite so that there is no possibility of confusion. It appears that the RTs (peaks) of the 5A and 5B in the F3 were too close together, as well as the etio and andro in the F2.
The only reference material we can use for all of the metabolites is the GCMS Cal Mix.
Do you agree?
Larry,
One last thing. Testosterone is endogenous to urine, all 4 of the metabolites to be tested for occur naturally in the blank urine. That's why I suggest it can be used as a reference material. Spiking it would just add more of those metabolites. OMJ made this point on DP.
M -
I think we now have the definitive statement of USADA's final position on how FL's S17 IRMS peaks were identified by LNDD. You and I, and everyone else who is interested, needs to go to p. 46, Section H, at the following link:
USADA's Final Word
This link is to USADA's proposed findings of fact and conclusions of law. I need to read this more carefully, but it appears that:
1. FL's S17 IRMS peaks were identified at LNDD (a) by using RTs or RRTs to identify the IRMS peak representing the internal standard 5aA-AC, (b) then counting large IRMS peaks to the right of the internal standard peak and idenfifying each peak by reference to the large peaks identified in the GC/MS test.
2. So, probably the best way to characterize LNDD's criteria here is not to call it "pattern matching", but "large peak counting".
M, we can go on with this discussion a while longer if you like, but I have another idea. Why don't you and I take this opportunity to "rebrief" our respective positions, using the new material available here and all of the stuff we've learned over the past few weeks. Here's my proposal:
1. You and I would "stipulate" that the method used at LNDD to identify IRMS peaks is the one described in the USADA document cited above. We might agree in advance to a short statement of the facts in the case, so we wouldn't have to repeat that in our briefs.
2. Then each of us would write a relatively short brief setting forth our respective positions. The brief would be limited to whether the LNDD's IRMS identification of FL's S17 sample is an ISL departure sufficient to overturn the FL doping finding. We'd agree in advance to keep our briefs under a specified word limit.
3. I would propose to write my brief here on TBV, in stages, so that everyone could comment. I'd reserve the right to make the final edits.
4. We could limit these briefs to whether there was an ISL violation. This way, we wouldn't have to address the burden shifting kinds of arguments we'd have to make on causation, should the ISL departure be established. We could save THAT argument for later.
5. We'd each write out briefs in as non-formal way as our training will allow. We'd keep the tone friendly and respectful.
What do you think? If you don't want to do this, I understand. You've taken on a tough enough job here, as is.
Regarding your 3:52 PM post, I think I agree with most of it. In theory, it should be possible to use the mix cal acetate to identify the chromatographic standard 5aA-AC in the FL fractions, but I have to look at this more closely. If the RTs match within 1%, then this sounds like a good method to me. As far as computing RTs from the chromatographic standard on both the GC/MS and the GC/IRMS and then comparing these times using an RRT analysis -- isn't that what Dr. M-A did and what Dr. Brenna and the majority decision said was invalid? If you can use this method to identify IRMS peaks within a 1% standard, that sounds like something that Dr. M-A would approve, but again I'd have to look at it more carefully. And yes, it might be useful to use the mix cal acetate to identify the 5bA and ketio -- those aren't the metabolites that were used to prove doping, but it still might be a useful technique. If it works, it might be nice to know that SOMETHING can be identified in the FL IRMS fractions. I think that's something that TBV tried to do here a while back.
Finally, yes I understand that the blank urine should contain all 4 testosterone metabolites of interest to us in doing the IRMS testing. It's just that the blank urine is not designed to be a reference substance. It's not the kind of "spiked" substance that TD2003IDCR requires (IMHO). I think you said the same thing in your 3:26 PM post.
M -
I need to take issue with one aspect of how you've described the identification criteria specified in TD2003IDCR. You've correctly pointed out that the TD2003IDCR criteria require RT of an analyte to differ by not more than 1% from that of the same substance in a reference sample. (Actually, the standard is 1% or 12 seconds, whichever is less.) But you've also said that the TD permits use of a more relaxed standard. So, for example, you argue in paragraph 2 of your updated post that "RTs must match within 1% although this limit can be relaxed", and in paragraph 7 you argue that "the relative retention times of the GCMS matched those in the GC-IRMS in compliance with TD2003IDCR when we relax the 1% matching requirement."
M, TD2003IDCR expressly permits the 1% (or 12 second) standard to be relaxed ONLY "in those cases where shifts in retention can be explained." USADA offered no explanation for shifts in retention, so there's no basis to relax the 1% (or 12 second) standard in this case.
TD2003IDCR offers one example of how a shift in retention can be explained: by "sample overload." Sample overload is where large amounts of sample are analyzed with the result that the GC capillary becomes saturated, so the retaining material in the capillary can no longer separate out the compounds in the way it is designed to do. Or in the language of the science: "The analytes with the highest affinity for the RP material will displace analystes of lower affinity." Sample Overload Explanationn
The sample overload example is instructive. TD2003IDCR is saying that the 1% (or 12 second) standard can be relaxed only when you can point to a specific explanation of why you weren't able to get the RTs to match up within normal limits. We don't have an explanation here to explain why there would have been a shift in retention times, other than the Duckworth explanation that LNDD did a lousy job setting up the chromatographic conditions in the GC/MS and the IRMS machines. That's not something that LNDD and USADA are likely to admit, nor would it be reasonable to read into TD2003IDCR that the 1% standard can be relaxed in cases where the lab doesn't do its job properly.
Moreover, even if you could provide an explanation for why the 1%/12 second standard should be relaxed in this case, I think you'd have to tell us what relaxed standard can be justified (2%? 3%? 5%?) based on your explanation. I don't think you can say, I have an explanation why the standard should be relaxed, therefore, the relaxed standard is whatever is necessary in order to validate the results obtained by LNDD. That would effectively eliminate the relaxed standard altogether. Luckily, neither one of us has to figure out what an appropriate relaxed standard might be, since USADA and LNDD never offered an explanation allowing the standard to be relaxed.
Again, look at the Brenna testimony. Brenna didn't say that there was sample overload or any particular reason why RTs would not match up in this case within the 1%/12 second criteria. Brenna said that it was simply impossible to match RTs or even RRTs between the GC/MS and IRMS machines. Brenna's testimony does not amount to an explanation for a shift in retention times that would justify a relaxed standard; it amounts to a confession that RRTs cannot be used in this case regardless of the standard.
So, M, at this point, if you want to argue that LNDD complied with the identification criteria set forth in TD2003IDCR, you'll have to live with the "unrelaxed" 1%/12 second standard. You have no basis on which to "relax"!
Two things:
First, M wrote:
"I tend to agree that the blank urine shouldn't be used as a GC-IRMS RT reference material (spiked urine sample) for the same reason I don't think that the Cal Mix Acetate even if it had contained all the metabolites should or could be used as an IRMS reference sample. Since there are no mass spectra, if you include all the metabolites, some of them are so close together that you can't be sure if you've identified them correctly by retention times alone in your Cal Mix."
Sure you can sure, as long as the chromatographic conditions are the same. Isn't that the way it's supposed to be done (or at least one of the ways)?
Second, before you guys do a whole lot more work, can one of you step back and remind me why you are arguing this out? TbV sort of started this discussion by saying that there was a violation of TD2003IDCR. Are you still arguing that fairly narrow (although by no means unimportant) legal issue, trying to decide whether there should have been a burden flip or not?
OR, Larry, in trying to show the unreliability of the IRMS peak identification, are you suggesting the (stronger) point that the peaks themselves really are misidentified? As in, "There is no 5A in the one labeled 5A, or 5B in the one labeled 5B" ?
OR, am I missing something altogether and this is really about the possibility of CONTAMINATION of the peaks.
Ms favorite DP poster, OMJ, has called this peak identification argument a red herring. And although I don't ultimately agree with OMJ, I think he's right about this (unless it is a matter of establishing the burden flip, as TbV originally suggested). The real issue is contamination of the peak(s), isn't it?
syi
Mike -
I come to the FL case from my perspective as a lawyer. I believe the arbitrators came to the wrong decision as a matter of law. Under my reading of the applicable rules, the arbitrators should have overturned the doping finding. I'm here to see if I can articulate the reasons why I've come to this conclusion in a manner that will persuade others. So far, I think I've achieved a very limited success, but I hope to do better.
You asked me what I'm trying to prove, using science-y kinds of terms. Am I trying to prove that the peaks are misidentified? Or that they're contaminated? Mike, you give me way, way too much credit. I'm not trying to prove anything that specific. I'm arguing that the WADA rules don't support the doping finding. If the WADA rules are based on good science, then perhaps my arguments will have some scientific validity. If not, not.
Let me try to be more specific. If I can make my case by showing that LNDD did not identify IRMS peaks within ISL standards, then I'm satisfied that I've FAIRLY proven an ISL departure, regardless of how many scientists might believe that the peaks were correctly identified.
Maybe if I had a perfect knowledge of the science, I could go along with pattern matching or peak counting or whatever it is that Brenna and OMJ say is OK. Or maybe I'd think like Dr. M-A that you need a quantifiable standard. I'd feel worse about my lack of scientific knowledge if there appeared to be anything like a scientific consensus on this point (or for that matter, about ANY of the science points at issue in this case). So long as the scientists continue to point in every different direction, I'll continue to push for this case being decided by the rules.
I know that this may sound wrong to you, or to others. We want the case to turn on the TRUTH. We think that the science is the key to the truth. But at least in the absence of any scientific consensus, my position is that the rules are the best available guide to the truth.
I can give you about a hundred reasons why I feel this way. Maybe the best reason is that the WADA rules are there for a reason. If USADA cannot prove its case against FL under the WADA rules, which aren't exactly athlete-friendly in the first place, then I'm confident that there's something substantively and scientifically wrong with their case against FL.
If the RRTs on the peaks are off by more than 1%, maybe that means that the peaks were misidentified, or maybe the peaks shifted position because they were contaminated, or maybe the peaks shifted because the chromotographic conditions were so terrible that the results of the test should not be trusted, or maybe the peaks shifted because the test was fudged in some way. I don't know, but I figure there's a reason why it happened and that the reason must be significant, even if we'll never know the reason.
Hope this makes sense.
Hi,
I really appreciate the effort Larry and M are putting in to get across the RT/RRT identification issue, because it is emblematic of the problems of making a point under this rule system.
Like Mr. Idiot, that particular point seems important to me because of the potential for causing a burden flip, and I look forward to one last go round to make the case about it clearer. I doubt we'll all reach agreement (though we can hope), but few stones will be unturned.
At the same time, while I think the identification and possibility of swap are important, I'm not sure it's going to be the dispositive issue. I still lean personally more to the contaminated peak, overlap, and integration error issues as being more ultimately illuminating.
The digging people have done here to find ignored MUST language that may cover the specificity requirements seems very interesting to investigate, and I suggest we do so in yet another new post since we're at 38 comments here.
The docu-dump problems are fixed (cross-fingers).
TBV
Post a Comment